WINC
A chromosome-painting-based pipeline to infer local ancestry under limited source availability
Supervised by Dr. Montinaro (University of Bari + University of Tartu) & Prof. Pagani (University of Padua + University of Tartu)
Why is this project cool?
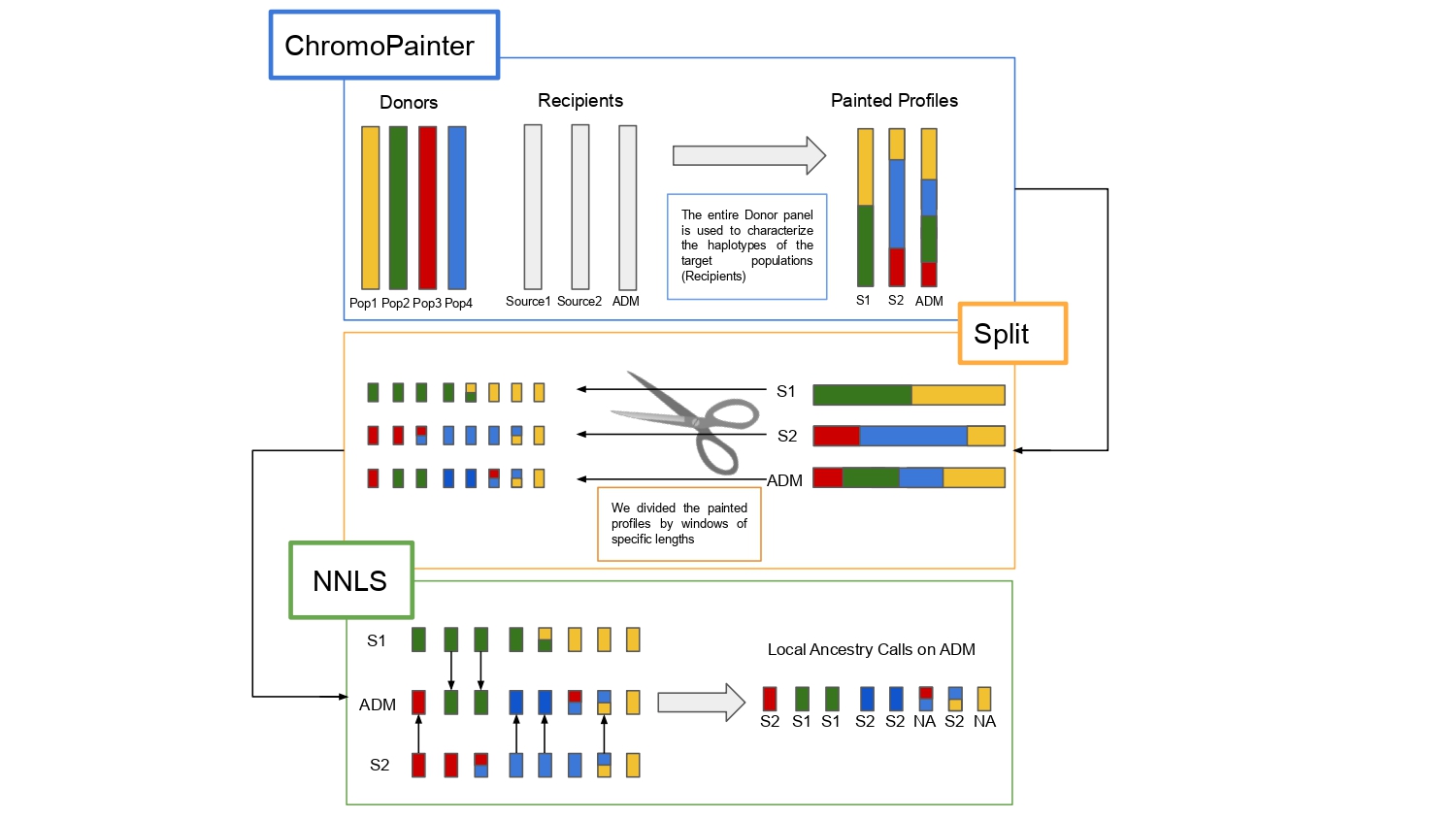
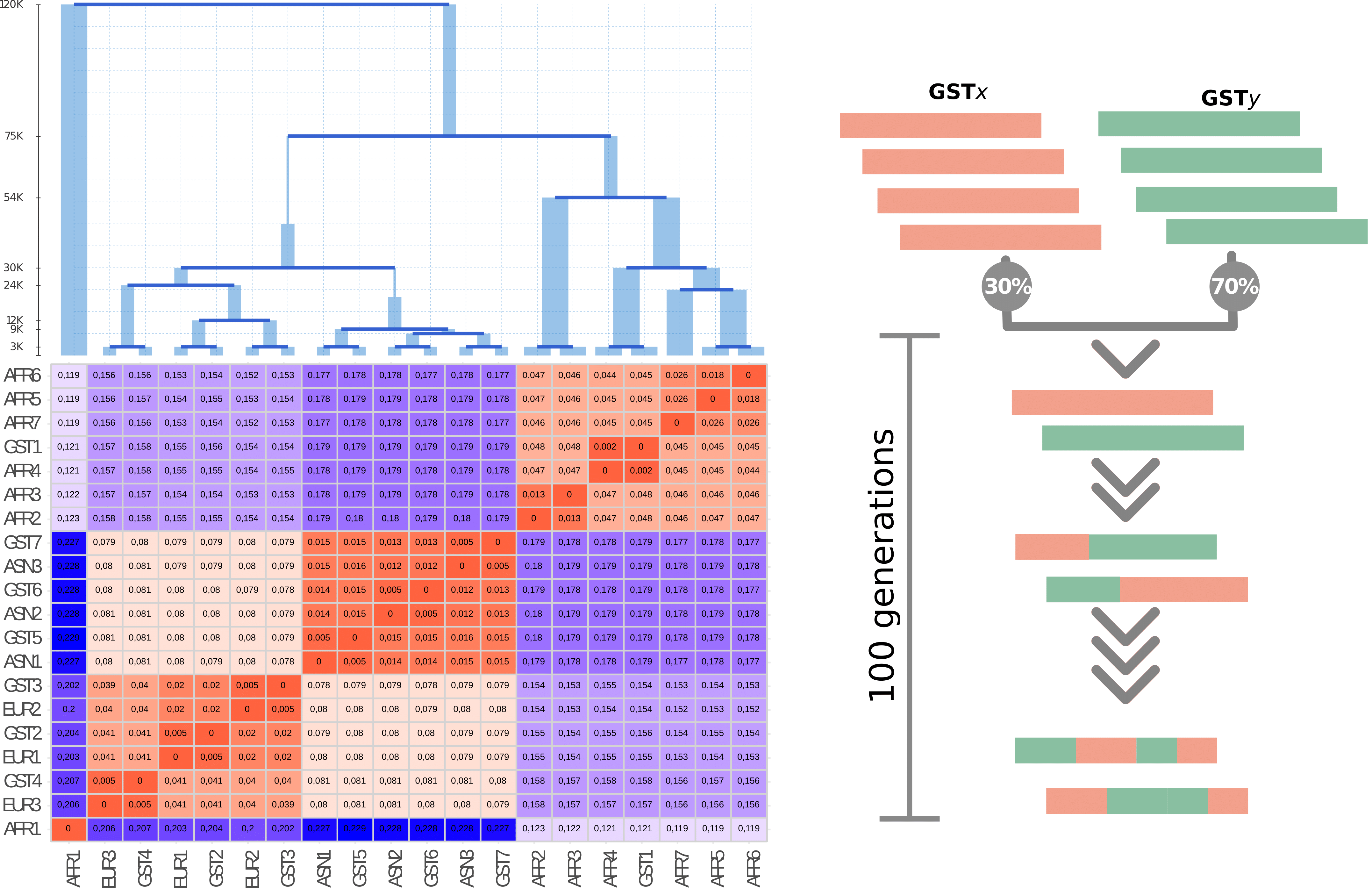
Contemporary individuals are the combination of genetic fragments inherited from ancestors belonging to multiple populations, as the result of migration and admixture. Isolating and characterizing these layers are crucial to the understanding of the genetic history of a given population. Ancestry deconvolution approaches make use of a large amount of source individuals, therefore constraining the performance of Local Ancestry Inferences when only few genomes are available from a given population. Here we present WINC, a local ancestry framework derived from the combination of ChromoPainter and NNLS approaches, as a method to retrieve local genetic assignments when only a few reference individuals are available. The framework is aided by a score assignment based on source differentiation to maximize the amount of sequences retrieved and is capable of retrieving accurate ancestry assignments when only two individuals for source populations are used.
References
2021
- A chromosome-painting-based pipeline to infer local ancestry under limited source availabilityGenome Biology and Evolution, 2021