Wild Boars
Hybridization of the Sardinian wild boar (Sus scrofa meridionalis)
Designed and managed by myself and Dr. Fabbri, under the supervision of Prof. Scandura (University of Sassari) + Prof. Pagani (University of Padua + University of Tartu)
Why is this project cool?
The wild boar (Sus scrofa meridionalis) arrived in Sardinia with the first human settlers in the early Neolithic. In this paper, we investigated the possible microevolutionary effects of the introgressive hybridization with domestic pig on the wild boar population, comparing Sardinian wild specimens with several commercial pig breeds and Sardinian local pigs, along with a putatively unadmixed wild boar population from Central Italy, all genotyped with a medium density SNP chip (~50K SNPs). Modelling the Sardinian boars as a mixture of a boar-like ancestry and a pig-like ancestry, we found three hybrids within the Sardinian boar samples out of 96 samples. The hybridization event was dated through MALDER around 20 generations ago (100 years ago).
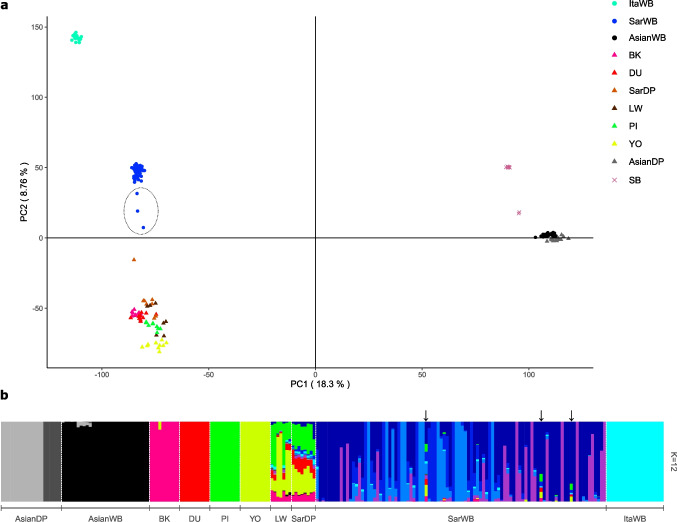
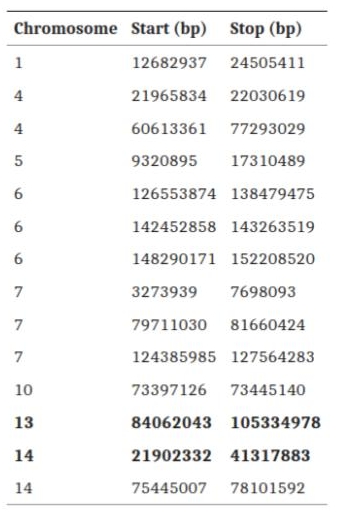
Local Ancestry Deconvolution analyses on the three hybrids revealed non-uniform levels of domestic pig introgression: ranging from 10% to 26.6%. Nonetheless, chromosomes 4, 7, 13, and 14 were the most affected, while chromosome 18 the least. Haplostrips, focused on the region of interest in chromosome 7, confirms the proportion of ancestry in the three hybrids: with one sample characterised with a domestic introgression and two with a wild boar-like haplotype.

We then applied selection analyses (PBS and XP-EHH) to further identify the putative genomic regions under selection. Due to the limited availability of SNPs as well as samples, we decided to opt for a randomization step. With this design we aimed to sample 3 non-hybrid Sardinian wild boars to use as an Outlier group and check how many and which windows would result significant just by chance with reduced non-hybrid sample size. We applied two randomization approaches, both of 1000 replications: a) 3 completely random SarWB b) 2 SarWB from eastern side of Sardinia + 1 from either north- or southwestern side (to account for population structure). All windows selected by the randomization analyses were further removed if found in the hybrid analyses. Selection analyses found two significant windows under selection enriched for the domestic alleles.
In conclusion, the hybridization with the domestic pig is not pervasive in the Sardinian population of wild boar: we identified three hybrids out of 96 samples (~3%). The domestic genes that seem to confer adaptive advantage in the introgressed pool are related to reproduction (P2RY214 and MED12L).
References
2023
- Anthropogenic hybridization and its influence on the adaptive potential of the Sardinian wild boar (Sus scrofa meridionalis)Journal of Applied Genetics, 2023